ProteoClade: A taxonomic toolkit for multi-species and metaproteomic analysis | PLOS Computational Biology

Python script to recode an amino acid sequence into generic format for... | Download Scientific Diagram

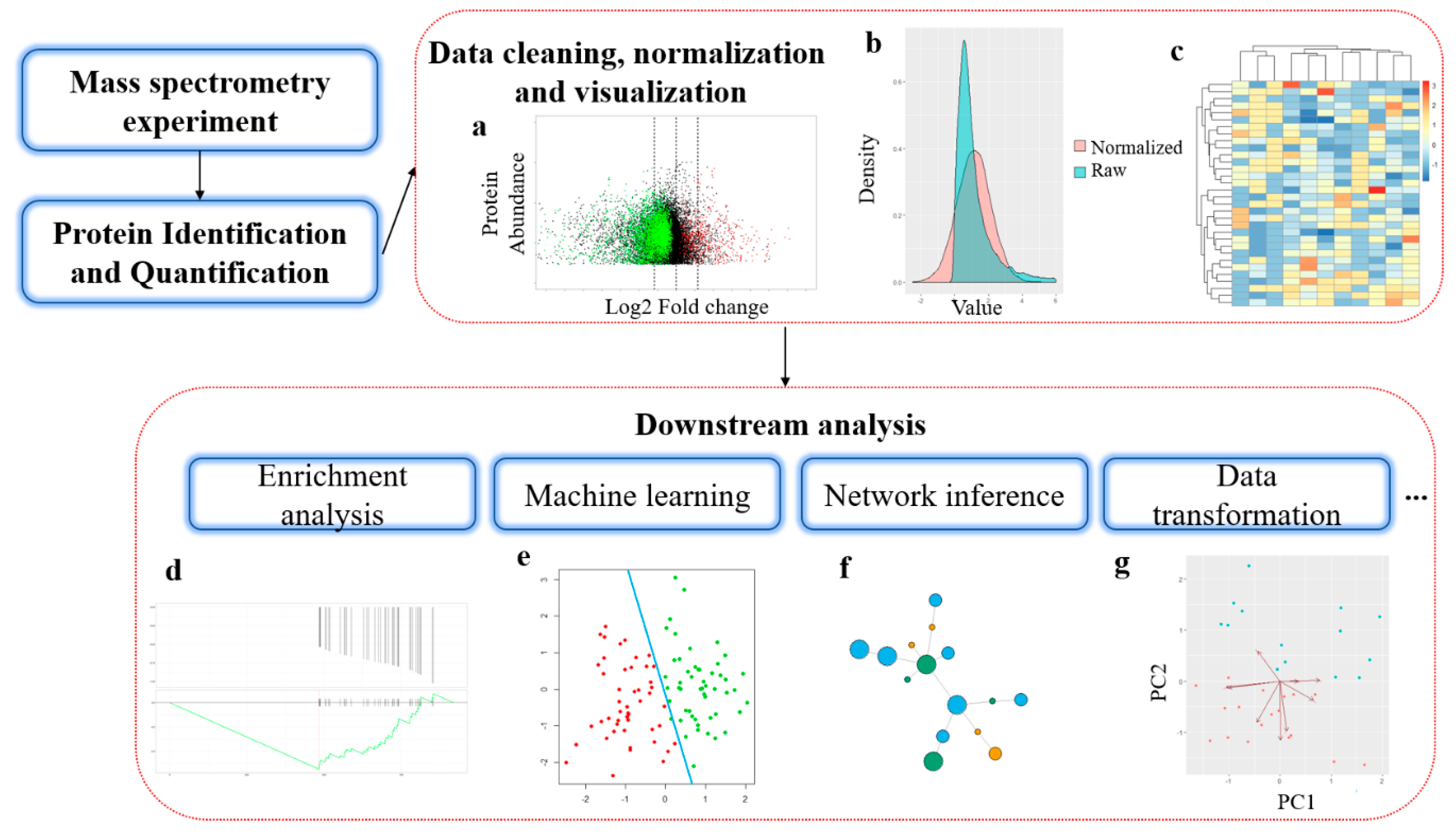
Data acquisition and database search. For a given experimental MS/MS... | Download Scientific Diagram
Functional expression of diverse post-translational peptide-modifying enzymes in Escherichia coli under uniform expression and purification conditions | PLOS ONE

Mutation Maker, An Open Source Oligo Design Platform for Protein Engineering | ACS Synthetic Biology

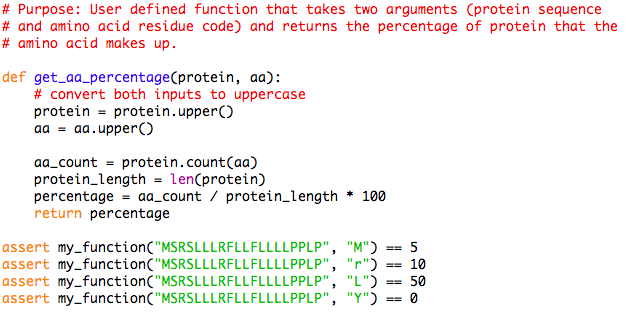
Molecules | Free Full-Text | PepFun: Open Source Protocols for Peptide-Related Computational Analysis
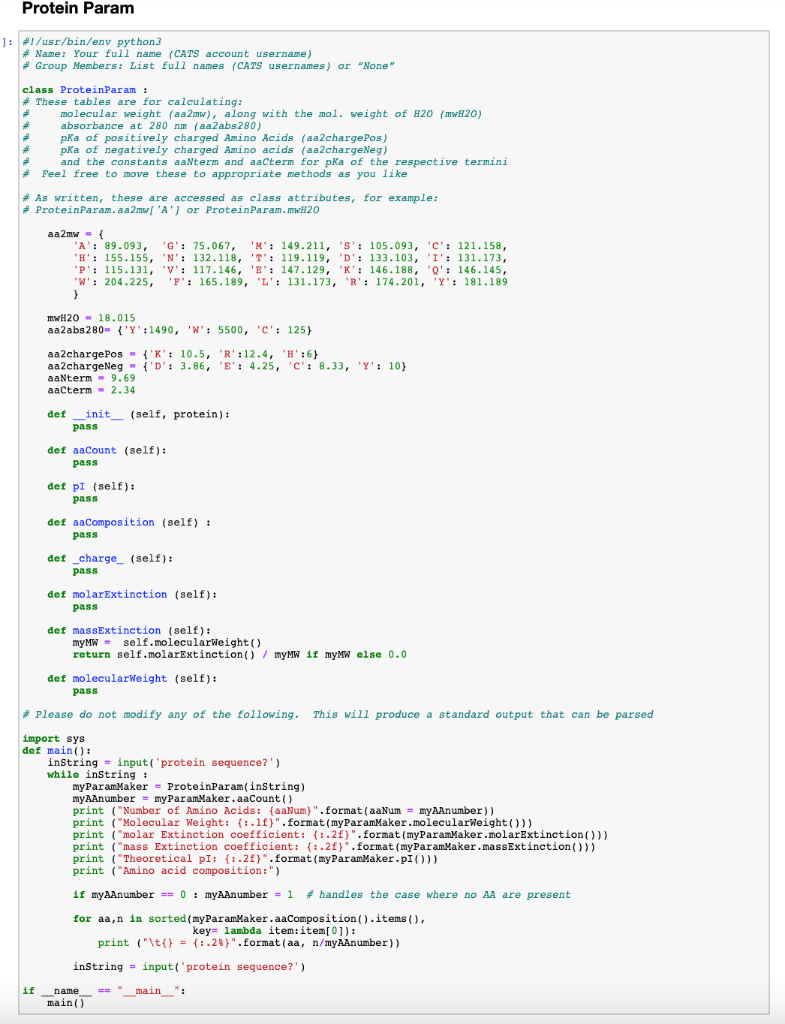
BioPython_Scripts/HowTo_Translate_DNA_fasta_to_Protein_Fasta.txt at master · PNNL-Comp-Mass-Spec/BioPython_Scripts · GitHub
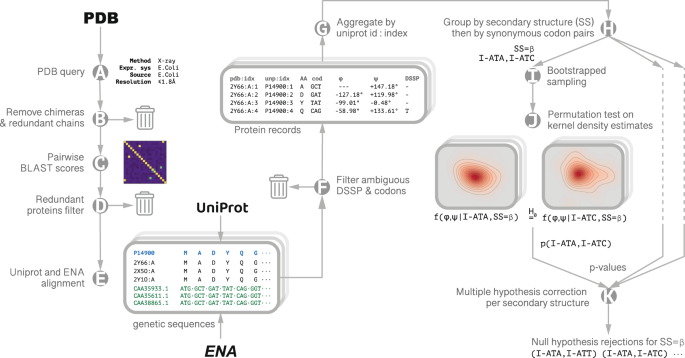
Codon-specific Ramachandran plots show amino acid backbone conformation depends on identity of the translated codon | Nature Communications
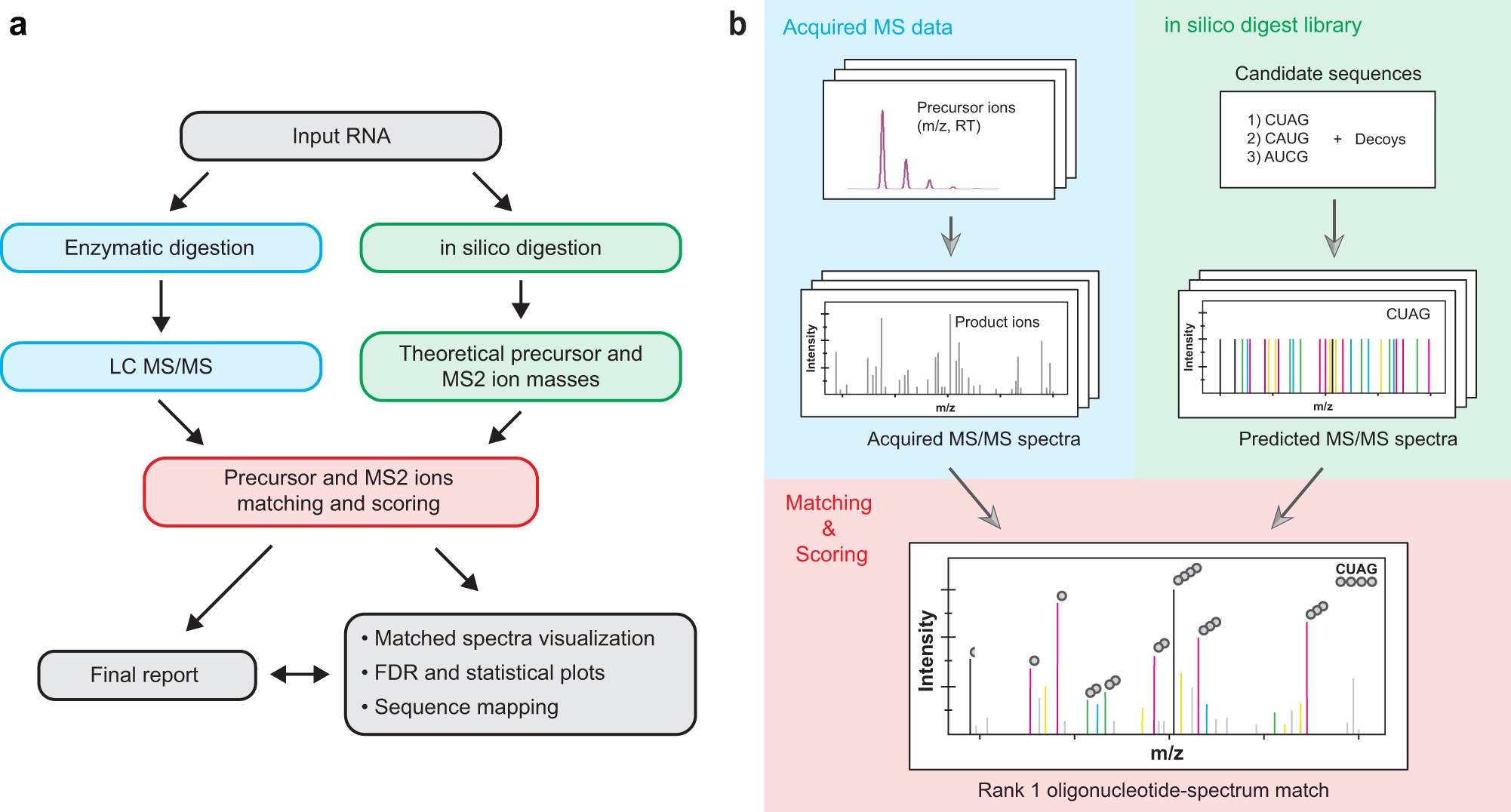
Pytheas: a software package for the automated analysis of RNA sequences and modifications via tandem mass spectrometry | Nature Communications

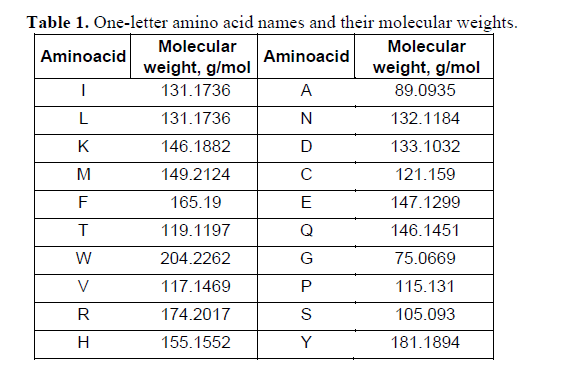
pIChemiSt ─ Free Tool for the Calculation of Isoelectric Points of Modified Peptides | Journal of Chemical Information and Modeling
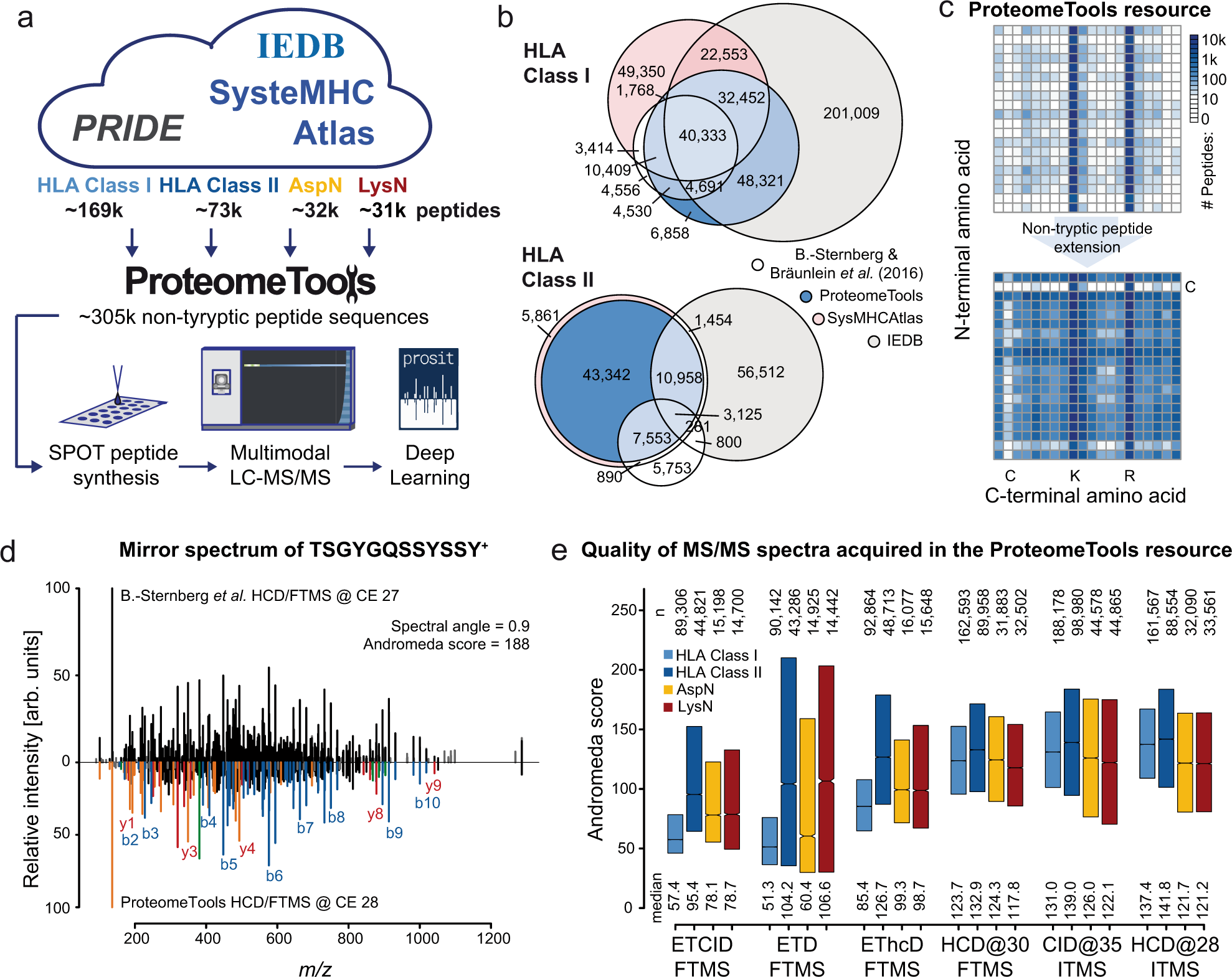
Deep learning boosts sensitivity of mass spectrometry-based immunopeptidomics | Nature Communications

Overview of the IPC 2.0 architecture. The input (amino acid sequence in... | Download Scientific Diagram